MAKER is one of the most popular bioinformatic pipeline used to annotate genomic information (Cantarel et al. 2008). MAKER utilizes standard programs in bioinformatics to customize the processing and preparation of the raw data. This includes processes to identify repeats, align ESTs and proteins to a target genomes, predict genes and quantify the quality of the results based on the provided evidence. MAKER focuses on automating the entire annotation process to create an easy and consistent initial annotation. MAKER is still under active development and is used in many areas of organism modeling.
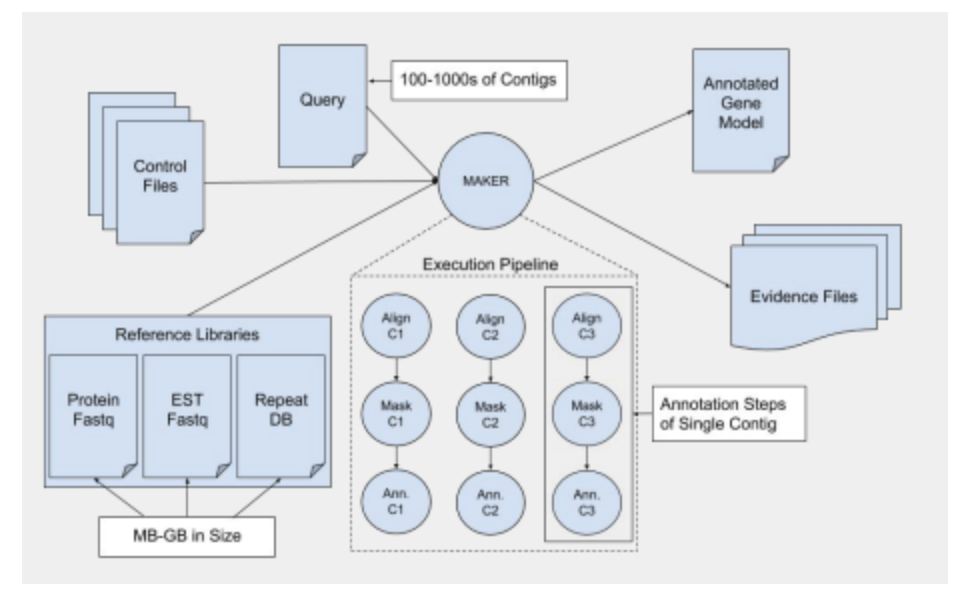
MAKER can be deployed as a sequential, multicore, or MPI application, depending on the available resources. As a side-effect of utilizing a number of different tools in the pipeline, files are often used as intermediaries on top of the data structures passed by MPI, leading to reliance on shared filesystems on multi-machine MPI runs
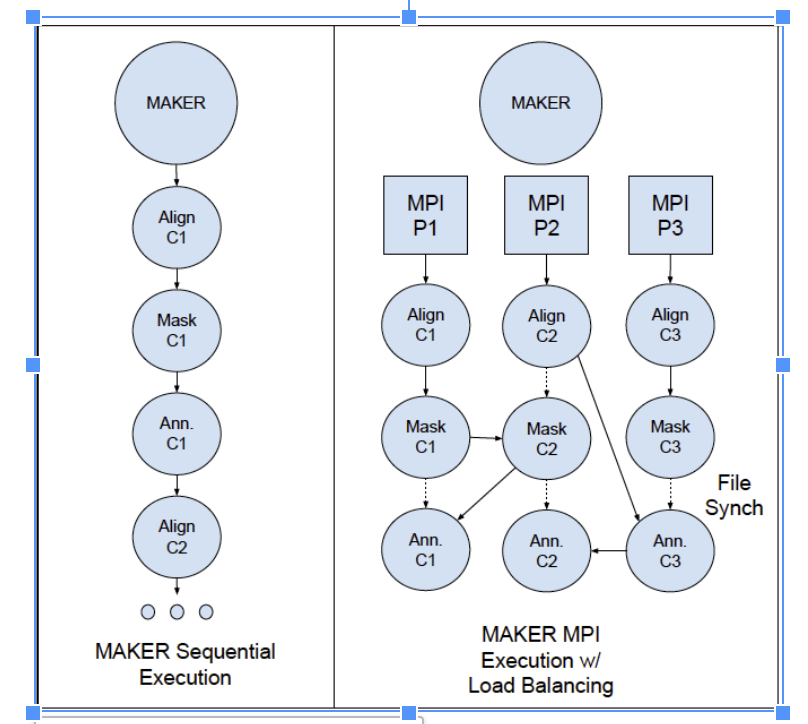
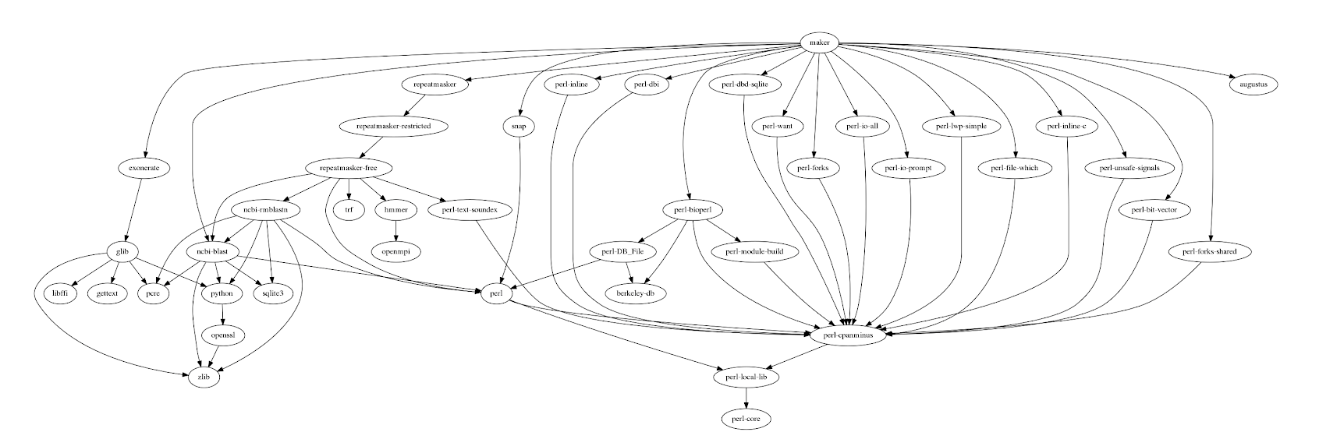
Although the scalability of MAKER makes it appropriate for projects of any size, deploying MAKER on traditional HPC clusters is widely seen as a strenuous task mainly because it has a large number of software dependencies that must be installed. MAKER itself is a amalgamation of many external bioinformatics tools, each with their own dependencies and hard-coded location for configuration files, making it difficult to configure it for a multi user setup typically found in HPC or any shared computational system. In addition, MAKER runs are not time efficient because they are performed sequentially from the input file (one sequence at a time).
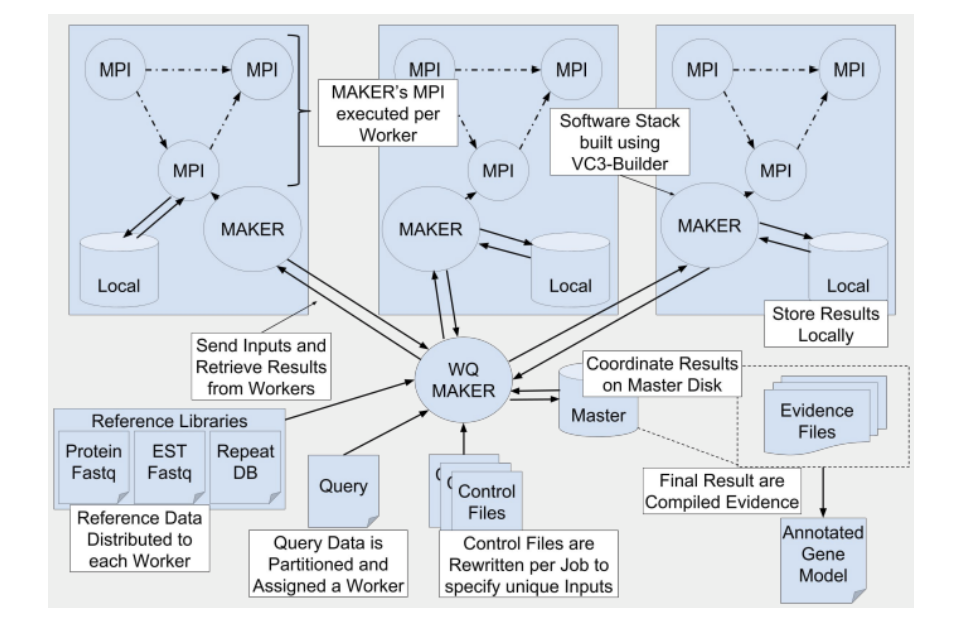
To overcome scalability and dependency problems with installing and managing MAKER, Thrasher et al. (2014) developed a version of MAKER with Workqueue (WQ-MAKER) designed to run on multiple virtual machines in the cloud (Thrasher et al. 2014). WQ-MAKER is built using the Work Queue API. The work is partitioned in different sizes, anywhere from individual sequences to the entire query file. WQ-MAKER does not split the work past the sequence level, as MAKER does with MPI, to prevent communication overhead from sub-processes. Each partition is a self-contained computational chunk that is distributed and organized after completion. WQ-MAKER utilizes Work Queue’s resource interface to allocate resources based on the partitions size and structure. Controlling at the task level allows for handling based on structure, such that long scaffolds are handled differently than short contigs. Using the resources allocated to a task by Work Queue, the worker can assign the appropriate amount of cores for MPI. To do this accurately assign resources a model is being developed as part of future work. Employing MPI on larger task, which occupy the entire worker, limits the master’s management burden of monitoring workers. In order to bring all these pieces together, we collaborated with Douglas Thain’s group at the University of Notre Dame to install WQ-MAKER application as a Jetstream image that uses coarse-grained parallelism over the network, and fine-grained parallelism (MPI or multi-core) within the single VM (Hazekamp et al. 2018). Because WQ-MAKER does not require any shared file system, this further reduces the file system load and allows for good scalability across commodity networks.
 Resources
Publication
Resources
Publication